5. Geospatial python#
Because of the wide array (pun intended) of open source data science modules that are accessible with python, it has emerged as a flagship language for processing, analyzing, and visualizing geospatial data. There are several key geospatial packages available in python:
Geopandas - processing vector data (e.g. shapfiles,
.GeoJSON,.KML, see here for a full list of geospatial vector filetypes)Rasterio - processing raster data (e.g. satellite imagery in
.tifor.tiffformat)Xarray - multidimensional array analysis (e.g. time series gridded or satellite datasets in
.ncor.hdfformat)GDAL - the key library (originally written in C and C++) that underscores most of the raster and vector processing operations in the above libraries, as well as GUI-based GIS software (ArcMap, QGIS, Google Earth/Maps, etc)
We will begin with (1) an (abridged) overview of map projections and coordinate systems, and (2) a brief discussion of the two fundamental types of geospatial data to motivate the topic.
Coordinate Systems and Projections#
A coordinate system is a framework used to determine position on the surface of the earth. Coordinate Systems are practical solutions to the challenge of depicting a 3D object (the earth) on a 2D surface (a screen).
Example of a coordinate system for a 2-D plane is the Cartesian Coordinate System (i.e. X and Y)
You’ve probably seen the Geographic Coordinate System (GCS), the three-dimensional coordinate system commonly used to define locations on the earth’s surface.
The unit of measure in the GCS is degrees,
locations are defined by latitude and longitude
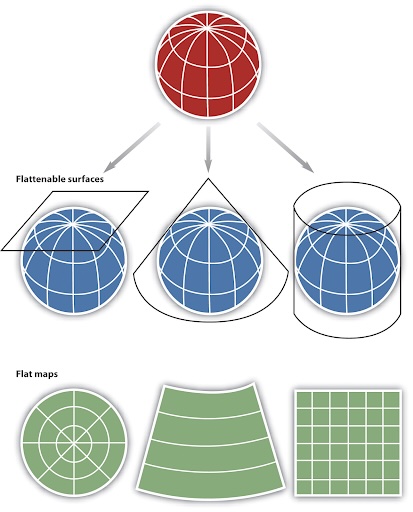
A Map Projection is how we transform geographic data from a three-dimensional, spherical coordinate system to a two-dimensional planar system. Map projections introduce distortions in distance, direction, and area. They are useful to convert data with units of degrees (in a GCS) into more practical length units such as meters or feet. There are several types of projected coordinate systems and projection types that can be used, the most common being:
Planar
Conical
Cylindrical
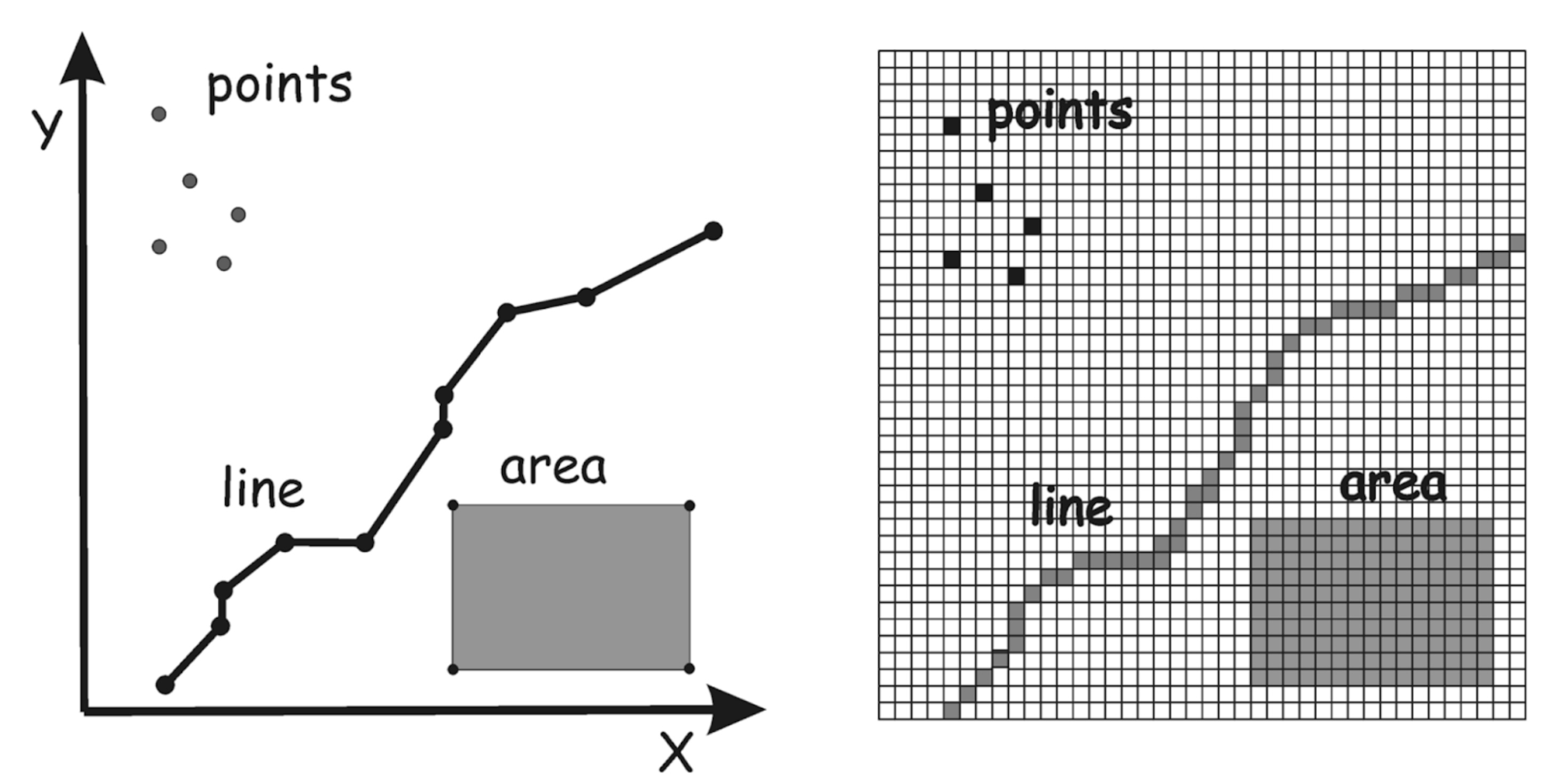
Vector and Raster data#
Geospatial data can be broadly categorized as either Vector or Raster:
Vector#
Vector data consit of points, lines, and polygons
Stored as tabular data files with geographic metadata
Lines are made up of points, have a starting and ending point
Polygons are made up of points, and the starting point = the ending point.
Can have a topology, or an organizational scheme to e.g. dictate the order or direction of data
Raster#
Raster data consist of grids of numbers - ‘Raster’ means ‘screen’ in Latin
Can contain multiple channels / bands - a true color image has Red, Green, Blue channels
The Spatial Resolution of a raster image refers to the size of a pixel (e.g. 1mm, 1cm, or 1m)
Satellite / drone / camera data is natively raster
The below image shows vector data on the left, and raster data on the right:
Now that we have some foundational Geospatial knowledge under our belt, let’s cover some of the python tools that we can use to process, analyze, and visualize geospatial data:
Geopandas#
Remember pandas from last module? Enter geopandas - a natural geospatial extension to store tabular vector data (think points, lines, and polygons) and simplify operations by keeping with pandas-like syntax.
Let’s load the same stations.csv file and convert it from a pandas.DataFrame to a geopandas.GeoDataFrame:
import pandas as pd
url = "https://raw.githubusercontent.com/py4wrds/py4wrds/refs/heads/module-4/data/gwl/stations.csv"
df = pd.read_csv(url)
df.head()
| STN_ID | SITE_CODE | SWN | WELL_NAME | LATITUDE | LONGITUDE | WLM_METHOD | WLM_ACC | BASIN_CODE | BASIN_NAME | COUNTY_NAME | WELL_DEPTH | WELL_USE | WELL_TYPE | WCR_NO | ZIP_CODE | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 51445 | 320000N1140000W001 | NaN | Bay Ridge | 35.5604 | -121.755 | USGS quad | Unknown | NaN | NaN | Monterey | NaN | Residential | Part of a nested/multi-completion well | NaN | 92154 |
| 1 | 25067 | 325450N1171061W001 | 19S02W05K003S | NaN | 32.5450 | -117.106 | Unknown | Unknown | 9-033 | Coastal Plain Of San Diego | San Diego | NaN | Unknown | Unknown | NaN | 92154 |
| 2 | 25068 | 325450N1171061W002 | 19S02W05K004S | NaN | 32.5450 | -117.106 | Unknown | Unknown | 9-033 | Coastal Plain Of San Diego | San Diego | NaN | Unknown | Unknown | NaN | 92154 |
| 3 | 39833 | 325450N1171061W003 | 19S02W05K005S | NaN | 32.5450 | -117.106 | Unknown | Unknown | 9-033 | Coastal Plain Of San Diego | San Diego | NaN | Unknown | Unknown | NaN | 92154 |
| 4 | 25069 | 325450N1171061W004 | 19S02W05K006S | NaN | 32.5450 | -117.106 | Unknown | Unknown | 9-033 | Coastal Plain Of San Diego | San Diego | NaN | Unknown | Unknown | NaN | 92154 |
# Create a GeoDataFrame from the DataFrame
import geopandas as gpd
gdf_stns = gpd.GeoDataFrame(
df,
geometry=gpd.points_from_xy(df["LONGITUDE"], df["LATITUDE"]),
crs="EPSG:4326", # Set the coordinate reference system (CRS) - more on this later
)
GeoDataFrame objects are a subclass of pandas.DataFrame, so you can use all of the same methods. In fact, a GeoDataFrame is just a pandas.DataFrame with a single extra column (geometry) and some extra features that make use of that column.
plot() is a special geopandas method that will plot the geometry / latlon of the df:
# Plot our results:
gdf_stns.plot()
<Axes: >
When printed, the tabular data is identical, and all pandas-like syntax can be used to sort and filter data:
# Get unique county names
print(gdf_stns["COUNTY_NAME"].unique())
# Plotting all stations in SLO county only
gdf_stns[gdf_stns["COUNTY_NAME"] == "San Luis Obispo"].plot()
['Monterey' 'San Diego' 'Imperial' 'Riverside' 'Orange' 'Kern'
'Los Angeles' 'San Bernardino' 'Ventura' 'Santa Barbara' 'Alameda'
'San Luis Obispo' 'Amador' 'Alpine' 'Tulare' 'Inyo' 'Fresno' 'Kings'
'Merced' 'San Benito' 'San Joaquin' 'Glenn' 'Madera' 'Santa Cruz'
'Santa Clara' 'Plumas' 'Stanislaus' 'San Mateo' 'Mono' 'Mariposa'
'San Francisco' 'Contra Costa' 'Tuolumne' 'Marin' 'Calaveras' 'Sonoma'
'Sacramento' 'Sutter' 'Solano' 'Napa' 'Placer' 'Yolo' 'Lake' 'Colusa'
'El Dorado' 'Mendocino' 'Yuba' 'Butte' 'Nevada' 'Sierra' 'Lassen'
'Tehama' 'Shasta' 'Humboldt' 'Modoc' 'Siskiyou' 'Del Norte' 'Klamath, OR']
<Axes: >
Reading and writing vector files#
Geopandas excels at making it easy to read and write vector files of several formats, which facilitates conversions and interoperability.
Simply import geopandas as gpd and then read any common vector file format with the gpd.read_file('filename') function, which works for common formats such as .shp (and supporting .dbf and .shx files), kml, kmz, geojson, etc..
# Read a geojson file
url = "https://raw.githubusercontent.com/scottpham/california-counties/refs/heads/master/caCountiesTopo.json"
gdf_counties = gpd.read_file(url)
gdf_counties.head()
| id | name | fullName | areaLand | geometry | |
|---|---|---|---|---|---|
| 0 | 06091 | Sierra | Sierra County | 2468686345 | POLYGON ((-120.14666 39.70738, -120.13527 39.7... |
| 1 | 06067 | Sacramento | Sacramento County | 2499176690 | POLYGON ((-121.14147 38.7118, -121.14043 38.71... |
| 2 | 06083 | Santa Barbara | Santa Barbara County | 7083926262 | MULTIPOLYGON (((-119.46758 34.06296, -119.4862... |
| 3 | 06009 | Calaveras | Calaveras County | 2641819811 | POLYGON ((-120.07212 38.50985, -120.07212 38.5... |
| 4 | 06111 | Ventura | Ventura County | 4773381212 | MULTIPOLYGON (((-119.63631 33.27314, -119.6363... |
gdf_counties.plot()
<Axes: >
To write files, simply use the syntax: gdf.to_file("path/to/out_file.geojson") syntax. We recommend storing files as .geojson due to readability and simplicity to store and retrieve.
gdf_counties.to_file("ca_counties.geojson")
/Users/andrew/opt/miniconda3/envs/py4wrds2/lib/python3.13/site-packages/pyogrio/geopandas.py:662: UserWarning: 'crs' was not provided. The output dataset will not have projection information defined and may not be usable in other systems.
write(
We received a warning that our CRS was not defined. We can eliminate this warning by defining the geographic WGS 84 coordinate reference system (CRS) through its EPSG code using gdf.set_crs('epsg:4236').
Similar to other DataFrame and pandas operations, we must specify the inplace = True argument:
gdf_counties.set_crs("epsg:4326", inplace=True)
gdf_counties.to_file("ca_counties.geojson")
# verify that file was written in our current directory:
import os
[x for x in os.listdir(os.getcwd()) if "geojson" in x]
['ca_counties.geojson']
We can inspect the coordinate reference system by using the gdf.crs syntax:
# Geographic CRS example:
gdf_stns.crs
<Geographic 2D CRS: EPSG:4326>
Name: WGS 84
Axis Info [ellipsoidal]:
- Lat[north]: Geodetic latitude (degree)
- Lon[east]: Geodetic longitude (degree)
Area of Use:
- name: World.
- bounds: (-180.0, -90.0, 180.0, 90.0)
Datum: World Geodetic System 1984 ensemble
- Ellipsoid: WGS 84
- Prime Meridian: Greenwich
Reprojections#
Often, we’ll want to change the coordinate system of some data. This can be achieved through reprojection.
UTM zones:#
Universal Transverse Mercator (UTM) is often an appropriate coordinate system for measuring distances, areas, etc
Units in meters, expressed in Easting / Northing
UTM coordinate systems
Let’s reproject the geodataframe into a projected CRS for California - UTM Zone 10 N, which is epsg:32611:
gdf_rpj = gdf_stns.to_crs("epsg:32611")
gdf_rpj.crs
<Projected CRS: EPSG:32611>
Name: WGS 84 / UTM zone 11N
Axis Info [cartesian]:
- E[east]: Easting (metre)
- N[north]: Northing (metre)
Area of Use:
- name: Between 120°W and 114°W, northern hemisphere between equator and 84°N, onshore and offshore. Canada - Alberta; British Columbia (BC); Northwest Territories (NWT); Nunavut. Mexico. United States (USA).
- bounds: (-120.0, 0.0, -114.0, 84.0)
Coordinate Operation:
- name: UTM zone 11N
- method: Transverse Mercator
Datum: World Geodetic System 1984 ensemble
- Ellipsoid: WGS 84
- Prime Meridian: Greenwich
the estimate_utm_crs() function may be convenient for going between geographic and projected CRS:
gdf_stns.estimate_utm_crs()
<Projected CRS: EPSG:32611>
Name: WGS 84 / UTM zone 11N
Axis Info [cartesian]:
- E[east]: Easting (metre)
- N[north]: Northing (metre)
Area of Use:
- name: Between 120°W and 114°W, northern hemisphere between equator and 84°N, onshore and offshore. Canada - Alberta; British Columbia (BC); Northwest Territories (NWT); Nunavut. Mexico. United States (USA).
- bounds: (-120.0, 0.0, -114.0, 84.0)
Coordinate Operation:
- name: UTM zone 11N
- method: Transverse Mercator
Datum: World Geodetic System 1984 ensemble
- Ellipsoid: WGS 84
- Prime Meridian: Greenwich
Geometric Operations#
We can easily calculate geometry properties such as area, centroids, bounds, and distances using geopandas.
Recall that one of our datasets consists of points (gdf_stns), and the other consists of polygons (gdf_counties)
Calculating Areas#
# Calculate area for each polygon in a UTM CRS
# Note - the units depend on the coordinate system,
# Be sure to reproject appropriately and perform conversions
gdf_rpj["area_sqkm"] = gdf_rpj.area * 1e-6 # Sq m to sq km
gdf_rpj.head()
| STN_ID | SITE_CODE | SWN | WELL_NAME | LATITUDE | LONGITUDE | WLM_METHOD | WLM_ACC | BASIN_CODE | BASIN_NAME | COUNTY_NAME | WELL_DEPTH | WELL_USE | WELL_TYPE | WCR_NO | ZIP_CODE | geometry | area_sqkm | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 51445 | 320000N1140000W001 | NaN | Bay Ridge | 35.5604 | -121.755 | USGS quad | Unknown | NaN | NaN | Monterey | NaN | Residential | Part of a nested/multi-completion well | NaN | 92154 | POINT (68916.162 3945609.103) | 0.0 |
| 1 | 25067 | 325450N1171061W001 | 19S02W05K003S | NaN | 32.5450 | -117.106 | Unknown | Unknown | 9-033 | Coastal Plain Of San Diego | San Diego | NaN | Unknown | Unknown | NaN | 92154 | POINT (490047.407 3600852.39) | 0.0 |
| 2 | 25068 | 325450N1171061W002 | 19S02W05K004S | NaN | 32.5450 | -117.106 | Unknown | Unknown | 9-033 | Coastal Plain Of San Diego | San Diego | NaN | Unknown | Unknown | NaN | 92154 | POINT (490047.407 3600852.39) | 0.0 |
| 3 | 39833 | 325450N1171061W003 | 19S02W05K005S | NaN | 32.5450 | -117.106 | Unknown | Unknown | 9-033 | Coastal Plain Of San Diego | San Diego | NaN | Unknown | Unknown | NaN | 92154 | POINT (490047.407 3600852.39) | 0.0 |
| 4 | 25069 | 325450N1171061W004 | 19S02W05K006S | NaN | 32.5450 | -117.106 | Unknown | Unknown | 9-033 | Coastal Plain Of San Diego | San Diego | NaN | Unknown | Unknown | NaN | 92154 | POINT (490047.407 3600852.39) | 0.0 |
Centroids#
The centroid is the geographic center point of a polygon (i.e. the mean of all the points that make up its surface)
Calculating centroids can be done with the gdf.centroid function:
print(gdf_counties.centroid.head())
gdf_counties.centroid.plot()
0 POINT (-120.5159 39.5804)
1 POINT (-121.34431 38.44933)
2 POINT (-120.03075 34.53832)
3 POINT (-120.55404 38.20466)
4 POINT (-119.12598 34.35742)
dtype: geometry
/var/folders/df/w347rlh96tx42h1kl69_4r8w0000gn/T/ipykernel_73601/3389763898.py:1: UserWarning: Geometry is in a geographic CRS. Results from 'centroid' are likely incorrect. Use 'GeoSeries.to_crs()' to re-project geometries to a projected CRS before this operation.
print(gdf_counties.centroid.head())
/var/folders/df/w347rlh96tx42h1kl69_4r8w0000gn/T/ipykernel_73601/3389763898.py:2: UserWarning: Geometry is in a geographic CRS. Results from 'centroid' are likely incorrect. Use 'GeoSeries.to_crs()' to re-project geometries to a projected CRS before this operation.
gdf_counties.centroid.plot()
<Axes: >
the alpha keyword here gives a transparency to the plotted points with 0 being fully transparent and 1 being fully opaque
Boundaries#
Similarly, boundaries can be obtained with the gdf.boundary function:
# Get the boundary of each polygon
print(gdf_counties.boundary.head())
gdf_counties.boundary.plot(alpha=0.3)
0 LINESTRING (-120.14666 39.70738, -120.13527 39...
1 LINESTRING (-121.14147 38.7118, -121.14043 38....
2 MULTILINESTRING ((-119.46758 34.06296, -119.48...
3 LINESTRING (-120.07212 38.50985, -120.07212 38...
4 MULTILINESTRING ((-119.63631 33.27314, -119.63...
dtype: geometry
<Axes: >
Distances#
Similarly, distances between two points can be computed with the distance function:
Let’s calculate the distance between each of the stations in our dataset
gdf_stns.plot(markersize=1, alpha=0.01)
<Axes: >
# Reproject to UTM zone 11N
gdf_stns.to_crs(gdf_stns.estimate_utm_crs(), inplace=True)
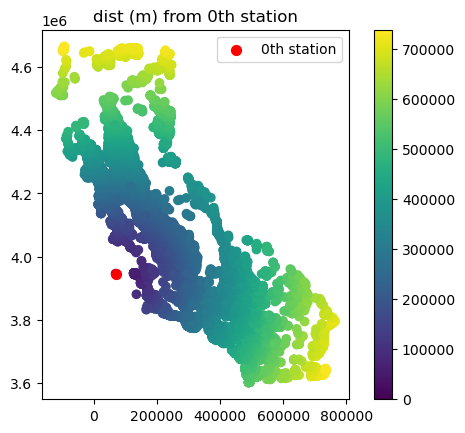
# calculate distance between the first station and each of the other stations
gdf_stns["dist_from_stn_1"] = gdf_stns.distance(gdf_stns.iloc[0].geometry)
# Plot stations, colored by distance
ax = gdf_stns.plot(column="dist_from_stn_1", legend=True)
# Plot the first station in red
gdf_stns.loc[[0], "geometry"].plot(markersize=50, color="red", ax=ax, label="0th station")
# add a legend a title
ax.legend()
ax.set_title("dist (m) from 0th station")
Text(0.5, 1.0, 'dist (m) from 0th station')
Plotting#
Just above we see an example of how to color a GeoDataFrame by a column, by specifying the column = 'column_name' argument into gdf.plot().
We can create interactive plots with the gdf.explore() funtion:
gdf_slo = gdf_stns[gdf_stns["COUNTY_NAME"] == "San Luis Obispo"]
gdf_slo.explore(legend=False)
Spatial Operations#
Geopandas makes it easy to perform basic spatial operations such as Buffers, Convex Hull, and Spatial Joins
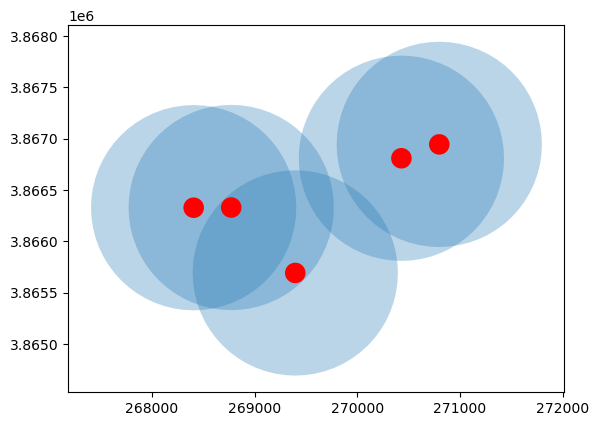
Buffer#
# Apply a 1000m buffer to the first 5 points in the SLO well data
ax = gdf_slo[:5].buffer(1000).plot(alpha=0.3)
# Plot the same 5 but with a smaller (100m) buffer
gdf_slo[:5].buffer(100).plot(ax=ax, color="red")
<Axes: >
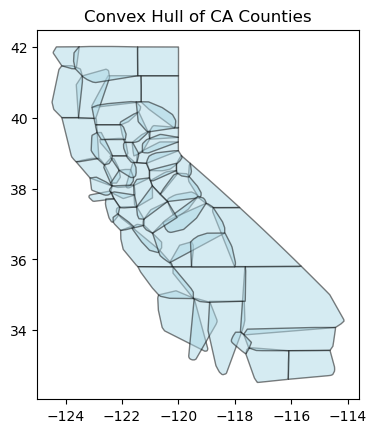
Convex Hull#
A convex hull gives the minimum bounds (as polygon) to enclose a number of points or another polygon’s points.
gdf_counties["convex_hull"] = gdf_counties.convex_hull
# Plot the convex hulls
ax = gdf_counties["convex_hull"].plot(alpha=0.5, color="lightblue", edgecolor="black")
ax.set_title("Convex Hull of CA Counties")
Text(0.5, 1.0, 'Convex Hull of CA Counties')
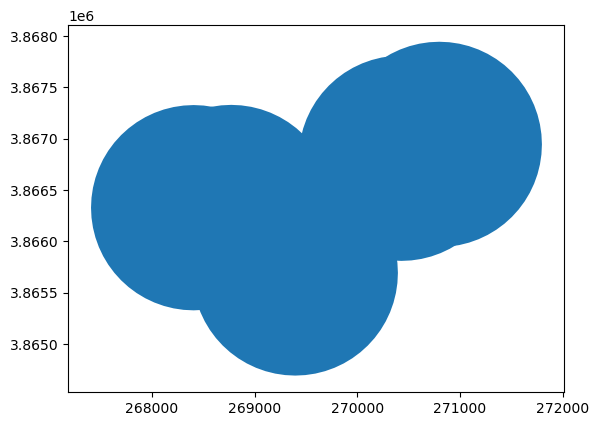
Dissolving Overlapping Polygons#
Say we want to create a new single polygon consisting of overlapping polygons, we can achieve this with a dissolve operation
# First we set the geometry column of the gdf to be the buffered polygons instead of the points
gdf_slo_subset = gdf_slo.iloc[:5, :]
gdf_slo_subset.loc[:, "geometry"] = gdf_slo_subset.buffer(1000)
gdf_slo_subset.plot()
<Axes: >
gdf_slo_subset.head()
| STN_ID | SITE_CODE | SWN | WELL_NAME | LATITUDE | LONGITUDE | WLM_METHOD | WLM_ACC | BASIN_CODE | BASIN_NAME | COUNTY_NAME | WELL_DEPTH | WELL_USE | WELL_TYPE | WCR_NO | ZIP_CODE | geometry | dist_from_stn_1 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 14732 | 14515 | 349075N1195236W001 | 10N25W35F001S | NaN | 34.9075 | -119.524 | Unknown | Unknown | 3-013 | Cuyama Valley | San Luis Obispo | NaN | Unknown | Unknown | NaN | 92154 | POLYGON ((270395.988 3865693.001, 270391.173 3... | 215821.092758 |
| 14834 | 32411 | 349130N1195350W001 | 10N25W27R001S | NaN | 34.9130 | -119.535 | Unknown | Unknown | 3-013 | Cuyama Valley | San Luis Obispo | NaN | Unknown | Unknown | NaN | 92154 | POLYGON ((269406.21 3866328.542, 269401.395 38... | 214666.454544 |
| 14835 | 38827 | 349131N1195309W001 | 10N25W27R002S | NaN | 34.9131 | -119.531 | Unknown | Unknown | 3-013 | Cuyama Valley | San Luis Obispo | NaN | Unknown | Unknown | NaN | 92154 | POLYGON ((269772.007 3866330.38, 269767.192 38... | 215005.755313 |
| 14924 | 14511 | 349178N1195134W001 | 10N25W25M003S | NaN | 34.9178 | -119.513 | Unknown | Unknown | 3-013 | Cuyama Valley | San Luis Obispo | NaN | Unknown | Unknown | NaN | 92154 | POLYGON ((271429.923 3866810.295, 271425.107 3... | 216372.474901 |
| 14942 | 32409 | 349191N1195092W001 | 10N25W25M004S | NaN | 34.9191 | -119.509 | Unknown | Unknown | 3-013 | Cuyama Valley | San Luis Obispo | NaN | Unknown | Unknown | NaN | 92154 | POLYGON ((271799.033 3866945.332, 271794.218 3... | 216667.216124 |
gdf_slo_subset.dissolve().plot()
<Axes: >
# Notice that there is only one row in the gdf after performing the dissolve
# values are inherited from the first row, but the geometry is the unary_union of all geometryes
gdf_slo_subset.dissolve().head()
| geometry | STN_ID | SITE_CODE | SWN | WELL_NAME | LATITUDE | LONGITUDE | WLM_METHOD | WLM_ACC | BASIN_CODE | BASIN_NAME | COUNTY_NAME | WELL_DEPTH | WELL_USE | WELL_TYPE | WCR_NO | ZIP_CODE | dist_from_stn_1 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | POLYGON ((270376.773 3865497.911, 270352.928 3... | 14515 | 349075N1195236W001 | 10N25W35F001S | None | 34.9075 | -119.524 | Unknown | Unknown | 3-013 | Cuyama Valley | San Luis Obispo | NaN | Unknown | Unknown | None | 92154 | 215821.092758 |
Spatial Joins, Queries, and Relations#
Intersections#
To check for spatial intersection between two geographic datasets, we can use the intersection function:
# Check which wells intersect with SLO's geometry
gdf_slo = gdf_counties[gdf_counties["name"] == "San Luis Obispo"].to_crs(gdf_counties.estimate_utm_crs())
gdf_slo.plot()
<Axes: >
slo_co_stns = gpd.overlay(gdf_stns, gdf_slo, how="intersection")
slo_co_stns.plot()
<Axes: >
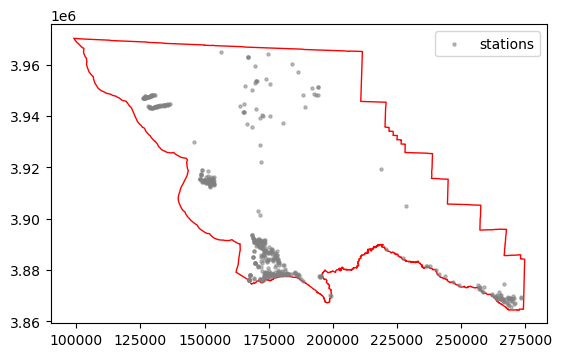
ax = gdf_slo.plot(color="white", edgecolor="red")
slo_co_stns.plot(alpha=0.5, markersize=5, color="gray", label="stations", ax=ax)
ax.legend()
<matplotlib.legend.Legend at 0x16c63cb90>
Exercise#
Determine how many “Industrial”, “Irrigation”, and “Observation” Wells are in SLO County
Create a basic map showing each category as a different color
Rasterio#
Rasterio is a powerful library for working with raster data (think: images or grids of numbers with pixels) written on top of the Geospatial Data Abstraction Library (GDAL), that makes it easy to perform a wide variety of raster processing and visualization tasks.
Reading and inspecting data#
We can open geospatial raster datasets (e.g. .tif and .tiff files) using the .open function:
import rasterio as rio
# raster_path = "https://github.com/opengeos/datasets/releases/download/raster/dem_90m.tif"
raster_path = "./data/dem_90m.tif"
src = rio.open(raster_path)
print(src)
<open DatasetReader name='./data/dem_90m.tif' mode='r'>
Notice that we are reading a file from a URL here, but this function works identically for local files.
Once a dataset has been opened, we can inspect lots of useful information, including:
Projection information
Bounding box
Spatial resolution
Dimensions - width / height
Other metadata
# Inspect crs
src.crs
CRS.from_wkt('PROJCS["WGS 84 / Pseudo-Mercator",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Mercator_1SP"],PARAMETER["central_meridian",0],PARAMETER["scale_factor",1],PARAMETER["false_easting",0],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],EXTENSION["PROJ4","+proj=merc +a=6378137 +b=6378137 +lat_ts=0 +lon_0=0 +x_0=0 +y_0=0 +k=1 +units=m +nadgrids=@null +wktext +no_defs"],AUTHORITY["EPSG","3857"]]')
# Get the EPSG code of the CRS
src.crs.to_epsg()
3857
# Inspect metadata
src.meta
{'driver': 'GTiff',
'dtype': 'int16',
'nodata': None,
'width': 4269,
'height': 3113,
'count': 1,
'crs': CRS.from_wkt('PROJCS["WGS 84 / Pseudo-Mercator",GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0,AUTHORITY["EPSG","8901"]],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AUTHORITY["EPSG","4326"]],PROJECTION["Mercator_1SP"],PARAMETER["central_meridian",0],PARAMETER["scale_factor",1],PARAMETER["false_easting",0],PARAMETER["false_northing",0],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH],EXTENSION["PROJ4","+proj=merc +a=6378137 +b=6378137 +lat_ts=0 +lon_0=0 +x_0=0 +y_0=0 +k=1 +units=m +nadgrids=@null +wktext +no_defs"],AUTHORITY["EPSG","3857"]]'),
'transform': Affine(90.0, 0.0, -13442488.3428,
0.0, -89.99579177642138, 4668371.5775)}
Discussion: What do the dtype, count, and nodata values in the src.meta dict mean?
# Inspect spatial res
src.res
(90.0, 89.99579177642138)
# Inspect dimensions or width / height
print(src.shape, src.width, src.height)
(3113, 4269) 4269 3113
# Inspect bounding box
src.bounds
BoundingBox(left=-13442488.3428, bottom=4388214.6777, right=-13058278.3428, top=4668371.5775)
Plotting and visualizing#
We can use the read function to read raster bands from .tiff or .tif files as numpy arrays, and then make use the imshow function from the trusty module matplotlib!
import numpy as np
import matplotlib.pyplot as plt
arr = src.read(1)
plt.imshow(arr)
plt.colorbar()
<matplotlib.colorbar.Colorbar at 0x1754dafd0>
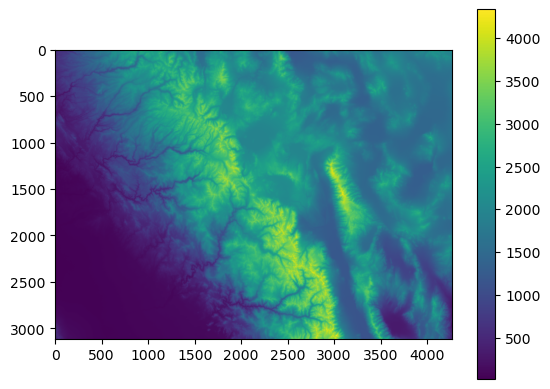
In the above code, we are reading the first band from the raster using src.read(1). For multiband rasters, we can read the other bands using the same syntax.
Note that this method does not preserve any projection information in the plot - the x and y axes are just the number of rows / cols in the dataset.
If we want to preserve any geospatial coordinate information, we can use the rasterio plotting module:
import rasterio.plot
rio.plot.show(src)
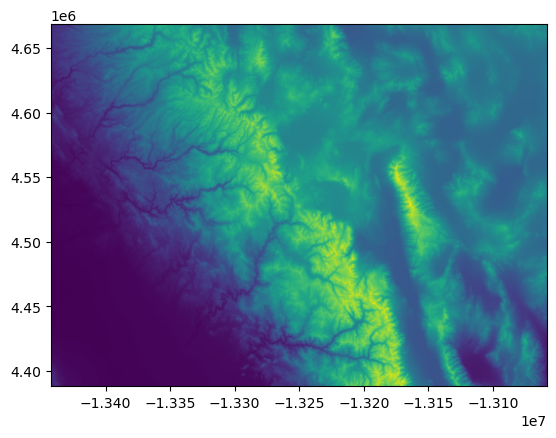
<Axes: >
Exercise#
set all values in raster less than 500 equal to np.nan
Set all values greater than 1000 to np.nan
Calculate the mean of the remaining raster cells
Hint - inspect the data type of the array using the
arr.dtypesyntax, and convert as needed
Formatting Plots#
We can leverage matplotlib’s functionality to style our plots, including adding titles and choosing colormaps:
fig, ax = plt.subplots(figsize=(8, 8))
rasterio.plot.show(src, cmap="terrain", ax=ax, title="Digital Elevation Model (DEM)")
plt.show()
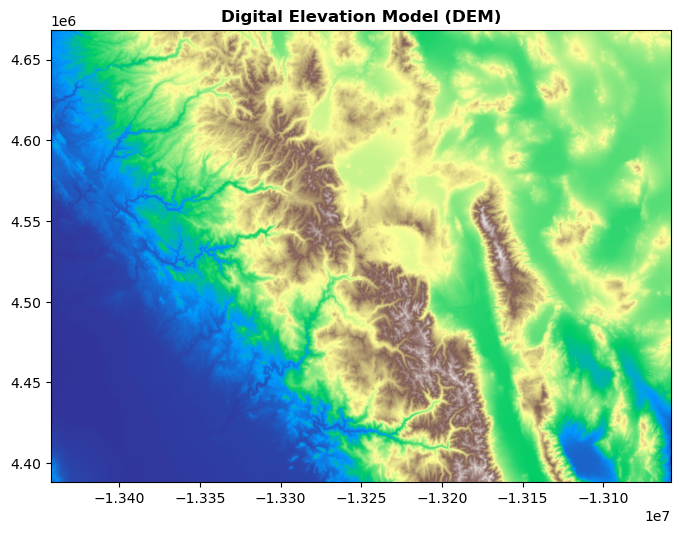
Plotting vector and raster data#
Tuolumne_co = gdf_counties[
gdf_counties["name"] == "Tuolumne"
].copy() # We need to make a copy of this object to avoid a SettingWithCopyWarning
Tuolumne_co.to_crs(src.crs, inplace=True)
fig, ax = plt.subplots(figsize=(8, 8))
rasterio.plot.show(src, cmap="terrain", ax=ax, title="Digital Elevation Model (DEM) and Tuolumne co")
Tuolumne_co.plot(ax=ax, edgecolor="red", facecolor="none", lw=2)
<Axes: title={'center': 'Digital Elevation Model (DEM) and Tuolumne co'}>
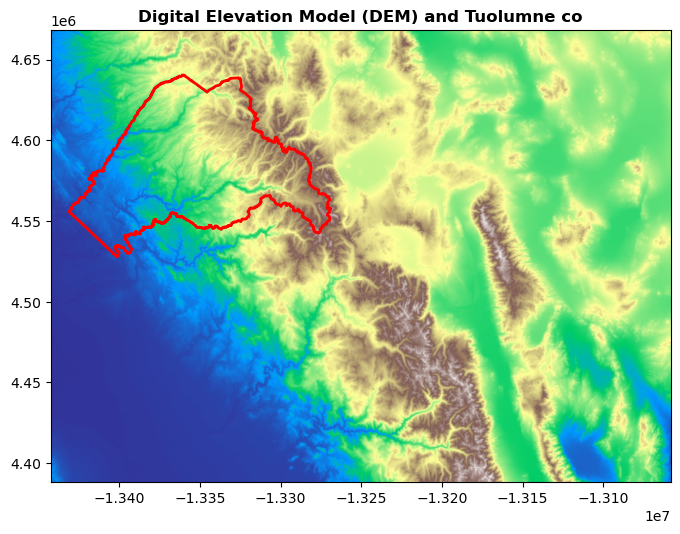
Clipping raster to vector data#
A common task involves clipping a raster to a vector dataset. We can use rasterio.mask to clip the DEM above to the boundary of Tuolumne county, defined in the ca_counties.geojson file:
import rasterio.mask
# Read raster
raster_path = "https://github.com/opengeos/datasets/releases/download/raster/dem_90m.tif"
raster_path = "./data/dem_90m.tif"
src = rio.open(raster_path)
# Read shape, isolate Tuolumne
shp_geom = gpd.read_file("ca_counties.geojson")
Tuolumne_co = shp_geom[shp_geom["name"] == "Tuolumne"].copy() # We need to make a copy of this object to avoid a SettingWithCopyWarning
# Ensure CRS are consistent
Tuolumne_co.to_crs(src.crs, inplace=True)
# Perform clip
out_image, out_transform = rio.mask.mask(src, Tuolumne_co["geometry"], crop=True)
# Verify results:
rasterio.plot.show(out_image[0, :, :]);
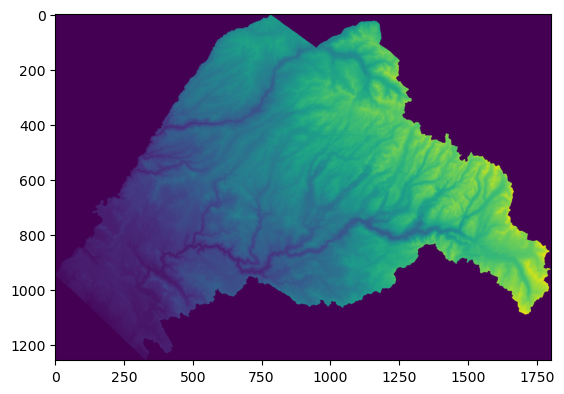
Notice that rasterio returns by default an array with 3 dimensions (out_image), for which we have selected the first band manually. This functionality exists to deal with multiband rasters.
Writing rasters#
Let’s save the raster we just clipped
# Set the output file
output_raster_path = "./data/clipped.tif"
# Extract some information from the image we processed
kwargs = src.meta
kwargs.update(height=out_image.shape[1], width=out_image.shape[2], count=out_image.shape[0], dtype=out_image.dtype, crs=src.crs)
# Save
with rasterio.open(output_raster_path, "w", **kwargs) as dst:
dst.write(out_image)
print(f"Raster data has been written to {output_raster_path}")
Raster data has been written to ./data/clipped.tif
Reprojecting Rasters#
We use the rasterio.warp module to change raster projections.
Let’s reproject our clipped raster to the WGS 84 (EPSG:3857) CRS and save the reprojected raster to a new file.
from rasterio.warp import calculate_default_transform, reproject, Resampling
# Set output filename
raster_path = "./data/clipped.tif"
dst_crs = "EPSG:4326" # WGS 84
output_reprojected_path = "./data/clipped_reprojected.tif"
with rasterio.open(raster_path) as src:
transform, width, height = calculate_default_transform(src.crs, dst_crs, src.width, src.height, *src.bounds)
profile = src.profile
profile.update(crs=dst_crs, transform=transform, width=width, height=height)
with rasterio.open(output_reprojected_path, "w", **profile) as dst:
for i in range(1, src.count + 1):
reproject(
source=rasterio.band(src, i),
destination=rasterio.band(dst, i),
src_transform=src.transform,
src_crs=src.crs,
dst_transform=transform,
dst_crs=dst_crs,
resampling=Resampling.nearest,
)
print(f"Reprojected raster saved at {output_reprojected_path}")
Reprojected raster saved at ./data/clipped_reprojected.tif
Multiband Rasters#
Satellite data is generally composed of a number of spectral bands. Each band measures the light reflected between two wavelengths (hence the term ‘band’). Rasterio allows us to work with these bands individually and in concert.
For example, NASA’s Landsat Satellites record the following bands:
Name |
Wavelength |
Description |
|---|---|---|
SR_B1 |
0.435-0.451 μm |
Band 1 (ultra blue, coastal aerosol) surface reflectance |
SR_B2 |
0.452-0.512 μm |
Band 2 (blue) surface reflectance |
SR_B3 |
0.533-0.590 μm |
Band 3 (green) surface reflectance |
SR_B4 |
0.636-0.673 μm |
Band 4 (red) surface reflectance |
SR_B5 |
0.851-0.879 μm |
Band 5 (near infrared) surface reflectance |
SR_B6 |
1.566-1.651 μm |
Band 6 (shortwave infrared 1) surface reflectance |
SR_B7 |
2.107-2.294 μm |
Band 7 (shortwave infrared 2) surface reflectance |
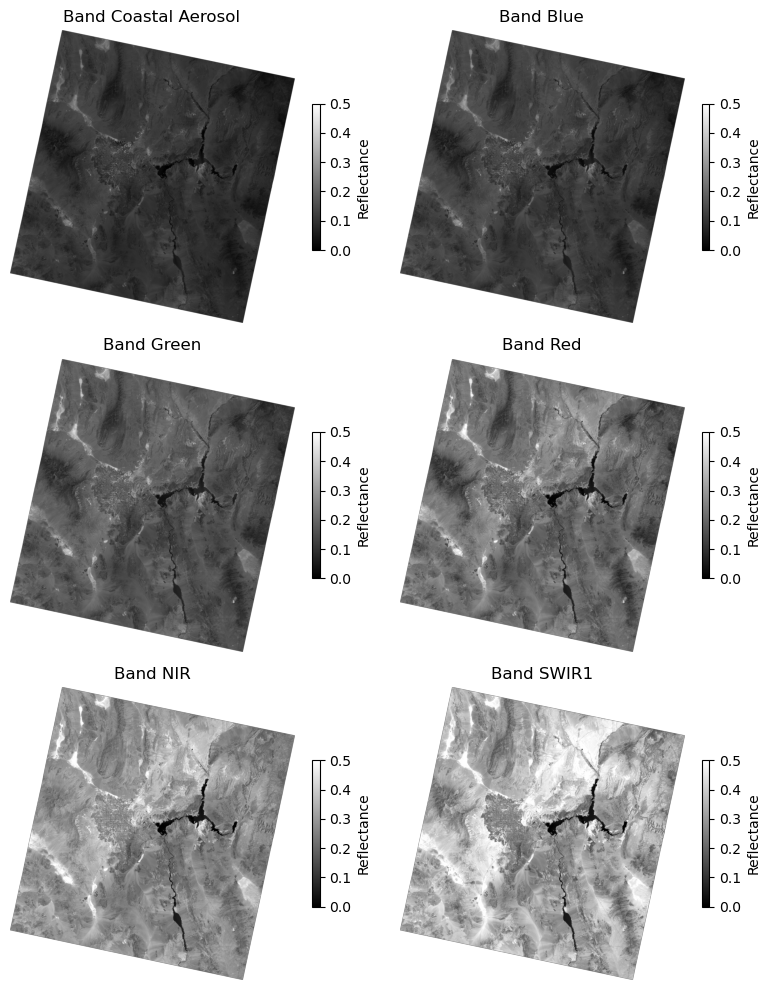
Let’s plot all the bands in this raster
# Read multiband Landsat image:
raster_path = "https://github.com/opengeos/datasets/releases/download/raster/LC09_039035_20240708_90m.tif"
# raster_path = "./data/LC09_039035_20240708_90m.tif"
src = rasterio.open(raster_path)
print(src)
# Specify band names:
band_names = ["Coastal Aerosol", "Blue", "Green", "Red", "NIR", "SWIR1", "SWIR2"]
<open DatasetReader name='https://github.com/opengeos/datasets/releases/download/raster/LC09_039035_20240708_90m.tif' mode='r'>
# Plot raster:
fig, axes = plt.subplots(nrows=3, ncols=2, figsize=(8, 10))
axes = axes.flatten() # Flatten the 2D array of axes to 1D for easy iteration
for band in range(1, src.count):
data = src.read(band)
ax = axes[band - 1]
im = ax.imshow(data, cmap="gray", vmin=0, vmax=0.5)
ax.set_title(f"Band {band_names[band - 1]}")
ax.axis("off")
fig.colorbar(im, ax=ax, label="Reflectance", shrink=0.5)
plt.tight_layout()
plt.show()
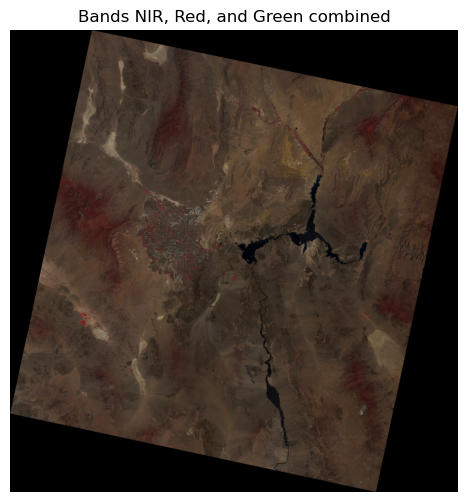
True color image#
In order to visualize a true color image, we need to visualize band 2 (blue), band 3 (green), and band 4 (red) in the same image:
nir_band = src.read(5)
red_band = src.read(4)
green_band = src.read(3)
# Stack the bands into a single array
rgb = np.dstack((nir_band, red_band, green_band)).clip(0, 1)
# Plot the stacked array
plt.figure(figsize=(6, 6))
plt.imshow(rgb)
plt.axis("off")
plt.title("Bands NIR, Red, and Green combined")
plt.show()
src.nodata
-inf
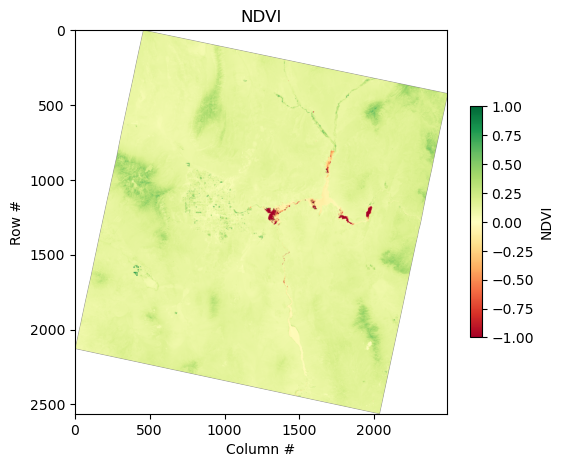
Band math: Calculating NDVI#
A common metric that we can derive from multiple bands is Normalized Difference Vegetation Index (NDVI), which is defined as:
# NDVI Calculation: NDVI = (NIR - Red) / (NIR + Red)
ndvi = (nir_band - red_band) / (nir_band + red_band)
/var/folders/df/w347rlh96tx42h1kl69_4r8w0000gn/T/ipykernel_73601/134765836.py:2: RuntimeWarning: invalid value encountered in subtract
ndvi = (nir_band - red_band) / (nir_band + red_band)
We received a RuntimeWarning about invalid values in our subtract operation. This is likely due to the ‘no data’ areas denoted by the black pixels in the true color image above. We can read the nodata value directly from rasterio and use numpy to change the nodata values in order to fix this issue:
# Set the nodata value to numpy nan for each of the bands
nir_band[nir_band == src.nodata] = np.nan
red_band[red_band == src.nodata] = np.nan
# NDVI Calculation: NDVI = (NIR - Red) / (NIR + Red)
ndvi = (nir_band - red_band) / (nir_band + red_band)
ndvi = ndvi.clip(-1, 1)
plt.figure(figsize=(6, 6))
plt.imshow(ndvi, cmap="RdYlGn", vmin=-1, vmax=1)
plt.colorbar(label="NDVI", shrink=0.5)
plt.title("NDVI")
plt.xlabel("Column #")
plt.ylabel("Row #")
plt.show()
Xarray#
Xarray makes working with labelled multi-dimensional arrays in Python simple, efficient, and fun!
Xarray extends NumPy functionality by providing data structures specifically for geospatial multi-dimensional arrays:
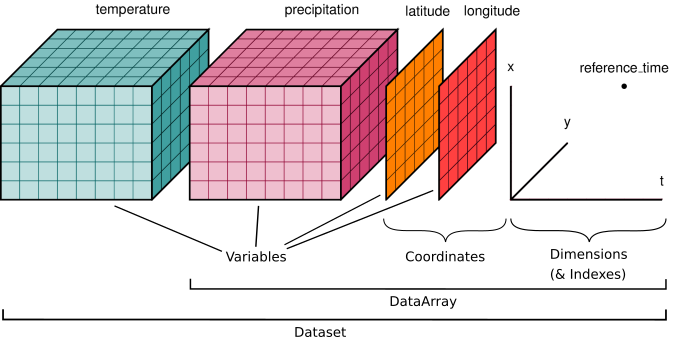
DataArray: A labeled, multi-dimensional array, which includes dimensions, coordinates, and attributes.
Dataset: A collection of DataArray objects that share the same dimensions.
Xarray data representation#
Xarray represents stacks of images as ‘cubes’, where two axes represent the latitude and longitude coordinates of spatial data, and a third axis represents time. Note that an xarray dataset can contain more than one variable!
The schematic below shows examples for temperature and precipitation
Loading Data and inspecting attributes#
This data was downloaded from NOAA: https://psl.noaa.gov/data/gridded/data.ncep.reanalysis.html
import xarray as xr
ds = xr.open_dataset("./data/runof.sfc.mon.mean.nc")
ds
<xarray.Dataset> Size: 67MB
Dimensions: (lat: 94, lon: 192, time: 923)
Coordinates:
* lat (lat) float32 376B 88.54 86.65 84.75 82.85 ... -84.75 -86.65 -88.54
* lon (lon) float32 768B 0.0 1.875 3.75 5.625 ... 352.5 354.4 356.2 358.1
* time (time) datetime64[ns] 7kB 1948-01-01 1948-02-01 ... 2024-11-01
Data variables:
runof (time, lat, lon) float32 67MB ...
Attributes:
description: Data is from NMC initialized reanalysis\n(4x/day). It co...
platform: Model
Conventions: COARDS
NCO: 20121013
history: Mon Jul 5 23:58:07 1999: ncrcat runof.mon.mean.nc /Datas...
title: monthly mean runof.sfc from the NCEP Reanalysis
dataset_title: NCEP-NCAR Reanalysis 1
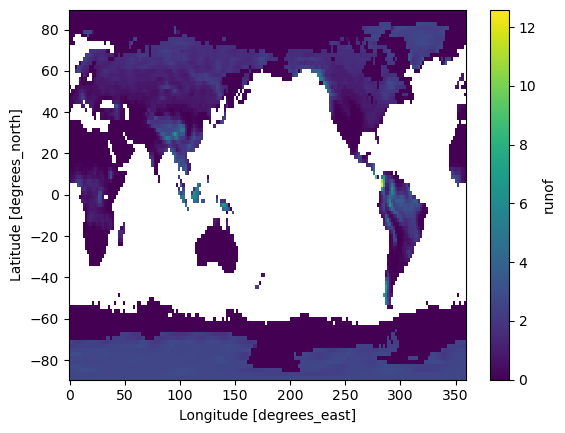
References: http://www.psl.noaa.gov/data/gridded/data.ncep.reanalysis...ds["runof"].mean(dim="time").plot()
<matplotlib.collections.QuadMesh at 0x38e041090>
# Select the data array describing runoff
ro = ds["runof"]
# Plot some of the data array attributes
# Obtain a numpy array of the values
print(ro.values.shape)
print(ro.values)
(923, 94, 192)
[[[ nan nan nan ... nan nan nan]
[ nan nan nan ... nan nan nan]
[ nan nan nan ... nan nan nan]
...
[1.1632297 1.1696817 1.1729066 ... 1.1600037 1.1600037 1.1600037]
[1.1600037 1.1600037 1.1600037 ... 1.1600037 1.1600037 1.1600037]
[1.1729066 1.1761327 1.1790358 ... 1.1696817 1.1696817 1.1729066]]
[[ nan nan nan ... nan nan nan]
[ nan nan nan ... nan nan nan]
[ nan nan nan ... nan nan nan]
...
[1.5599976 1.5599976 1.5599976 ... 1.4944816 1.5186188 1.5427573]
[1.5389636 1.5389636 1.5389636 ... 1.5358603 1.5358603 1.5389636]
[1.5599976 1.5599976 1.5599976 ... 1.5599976 1.5599976 1.5599976]]
[[ nan nan nan ... nan nan nan]
[ nan nan nan ... nan nan nan]
[ nan nan nan ... nan nan nan]
...
[1.5599976 1.5599976 1.5599976 ... 1.5599976 1.5599976 1.5599976]
[1.5599976 1.5599976 1.5599976 ... 1.5599976 1.5599976 1.5599976]
[1.5599976 1.5599976 1.5599976 ... 1.5599976 1.5599976 1.5599976]]
...
[[0. 0. 0. ... 0. 0. 0. ]
[0. 0. 0. ... 0. 0. 0. ]
[0. 0. 0. ... 0. 0. 0. ]
...
[2.8000002 2.9566665 3.1999993 ... 2.8000002 2.8000002 2.8000002]
[2.8000002 2.8000002 2.8000002 ... 2.8000002 2.8000002 2.8000002]
[2.8000002 2.8000002 2.8000002 ... 2.8000002 2.8000002 2.8000002]]
[[0. 0. 0. ... 0. 0. 0. ]
[0. 0. 0. ... 0. 0. 0. ]
[0. 0. 0. ... 0. 0. 0. ]
...
[2.8000004 2.8000004 3.0354831 ... 2.8000004 2.8000004 2.8000004]
[2.8000004 2.8000004 2.8000004 ... 2.8000004 2.8000004 2.8000004]
[2.8000004 2.8000004 2.8000004 ... 2.8000004 2.8000004 2.8000004]]
[[0. 0. 0. ... 0. 0. 0. ]
[0. 0. 0. ... 0. 0. 0. ]
[0. 0. 0. ... 0. 0. 0. ]
...
[2.8000002 2.8000002 2.8000002 ... 2.8000002 2.8000002 2.8000002]
[2.8000002 2.8000002 2.8000002 ... 2.8000002 2.8000002 2.8000002]
[2.8000002 2.8000002 2.8000002 ... 2.8000002 2.8000002 2.8000002]]]
print(ro.coords)
Coordinates:
* lat (lat) float32 376B 88.54 86.65 84.75 82.85 ... -84.75 -86.65 -88.54
* lon (lon) float32 768B 0.0 1.875 3.75 5.625 ... 352.5 354.4 356.2 358.1
* time (time) datetime64[ns] 7kB 1948-01-01 1948-02-01 ... 2024-11-01
print(ro.attrs)
{'long_name': 'Monthly Mean of Water Runoff', 'valid_range': array([-400., 700.], dtype=float32), 'units': 'kg/m^2', 'precision': np.int16(1), 'var_desc': 'Water Runoff', 'level_desc': 'Surface', 'statistic': 'Mean', 'parent_stat': 'Individual Obs', 'dataset': 'NCEP Reanalysis Derived Products', 'actual_range': array([-5.e+00, 1.e+30], dtype=float32)}
print(ro.dims)
('time', 'lat', 'lon')
Filtering and subsetting#
We can easily select data based on dimension labels, which is very intuitive when working with geospatial data.
# Select data for a specific time and location
selected_data = ro.sel(time="2000-01-01", lat=40.0, lon=120.0, method="nearest")
selected_data
<xarray.DataArray 'runof' ()> Size: 4B
array(1.587088, dtype=float32)
Coordinates:
lat float32 4B 40.95
lon float32 4B 120.0
time datetime64[ns] 8B 2000-01-01
Attributes:
long_name: Monthly Mean of Water Runoff
valid_range: [-400. 700.]
units: kg/m^2
precision: 1
var_desc: Water Runoff
level_desc: Surface
statistic: Mean
parent_stat: Individual Obs
dataset: NCEP Reanalysis Derived Products
actual_range: [-5.e+00 1.e+30]# Slice data across a range of times
time_slice = ro.sel(time=slice("2013-01-01", "2013-01-31"))
time_slice
<xarray.DataArray 'runof' (time: 1, lat: 94, lon: 192)> Size: 72kB
array([[[0. , 0. , ..., 0. , 0. ],
[0. , 0. , ..., 0. , 0. ],
...,
[2.580681, 2.590358, ..., 2.567777, 2.574229],
[2.800049, 2.800049, ..., 2.800049, 2.800049]]],
shape=(1, 94, 192), dtype=float32)
Coordinates:
* lat (lat) float32 376B 88.54 86.65 84.75 82.85 ... -84.75 -86.65 -88.54
* lon (lon) float32 768B 0.0 1.875 3.75 5.625 ... 352.5 354.4 356.2 358.1
* time (time) datetime64[ns] 8B 2013-01-01
Attributes:
long_name: Monthly Mean of Water Runoff
valid_range: [-400. 700.]
units: kg/m^2
precision: 1
var_desc: Water Runoff
level_desc: Surface
statistic: Mean
parent_stat: Individual Obs
dataset: NCEP Reanalysis Derived Products
actual_range: [-5.e+00 1.e+30]# Calculate the mean SWE over time
mean_ro = ro.mean(dim="time")
mean_ro
<xarray.DataArray 'runof' (lat: 94, lon: 192)> Size: 72kB
array([[0. , 0. , 0. , ..., 0. , 0. ,
0. ],
[0. , 0. , 0. , ..., 0. , 0. ,
0. ],
[0. , 0. , 0. , ..., 0. , 0. ,
0. ],
...,
[2.635003 , 2.6681736, 2.7444425, ..., 2.6152654, 2.6228926,
2.6288702],
[2.6165829, 2.6178818, 2.6189005, ..., 2.6109586, 2.6131048,
2.6150517],
[2.6433473, 2.6445358, 2.645743 , ..., 2.6407027, 2.641557 ,
2.6424491]], shape=(94, 192), dtype=float32)
Coordinates:
* lat (lat) float32 376B 88.54 86.65 84.75 82.85 ... -84.75 -86.65 -88.54
* lon (lon) float32 768B 0.0 1.875 3.75 5.625 ... 352.5 354.4 356.2 358.1Plotting and visualization#
Xarray provides builtin plotting methods that can be readily modified and integrated with standard matplotlib syntax
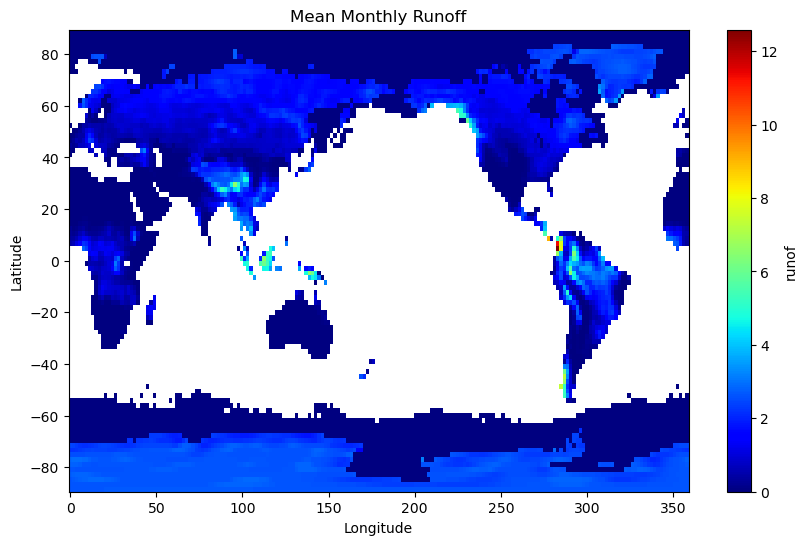
mean_ro.plot(cmap="jet", figsize=(10, 6))
plt.xlabel("Longitude")
plt.ylabel("Latitude")
plt.title("Mean Monthly Runoff")
Text(0.5, 1.0, 'Mean Monthly Runoff')
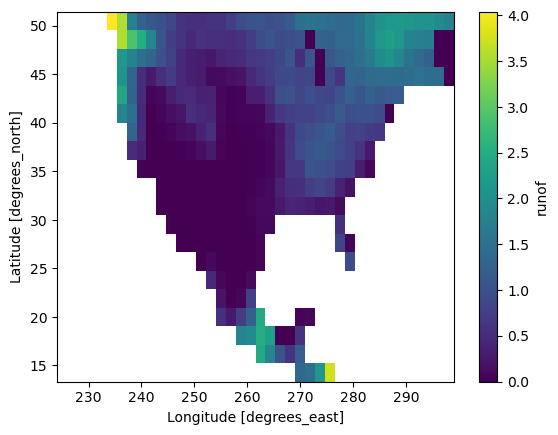
We can use the .isel method to select a spatial subset of the data by index:
cropped_ds = mean_ro.isel(lat=slice(20, 40), lon=slice(120, 160))
cropped_ds.plot()
<matplotlib.collections.QuadMesh at 0x38e70ad50>
# Plot a time series for a specific location
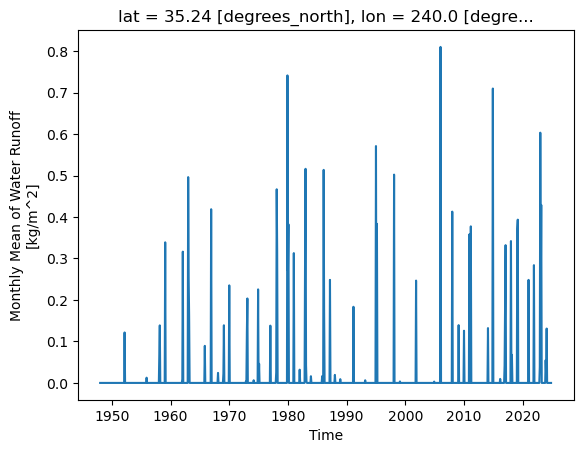
ro.sel(lat=35.0, lon=240, method="nearest").plot()
plt.show()
GDAL#
The Geospatial Data Abstraction Library (GDAL) is the magical piece of open source software that underlies all of the above libraries.
While GDAL has python bindings, the syntax are more complicated and less user friendly than the above libraries. Thus, coverage of GDAL syntax explicitly is outside the scope of this course.
For large batch, big data, or highly custom processing tasks, GDAL has a large array of command-line utilities (see here) that can be deployed via the Anaconda Prompt command line that was introduced in module 1.


